
Introduction
Dorado is Oxford Nanopore’s open source production basecaller. It converts raw signal data from the Nanopore sequencer into nucleotide sequences using a machine learning algorithm optimized for GPUs. The github claims linear scaling on GPUs clusters.
Here, we benchmark basecalling using the ResearchBox assistant on multi-GPU instances with 1×, 4×, and 8× Nvidia L4 GPUs to evaluate the “linear scaling” claim in practice. We can show the scaling for the super-accurate "sup" model is indeed close to perfectly linear.
Total time for you to reproduce and run this linear scaling benchmark on ResearchBox.ai : ~2 hours (scroll to end to see how)
Experiment setup
Dataset: This benchmark uses a POD5 dataset from Kirkegaard, R. H
Software: Benchmarks used the official Dorado v1.1 command-line binary. Dorado lets you choose model families tuned for different speed/accuracy trade-offs: fast, hac (high-accuracy), and sup (super-accurate). We used the sup model here. Dorado logs show the exact model used was dna_r10.4.1_e8.2_400bps_sup@v5.2.0. The model name tells us chemistry version and translocation speed too.
Compute: The ResearchBox.ai chat assistant was used to configure a nanopore basecalling workspace on a Nvidia L4 cloud instance. After running the benchmark, the assistant was asked to upsize the box to multi GPU instances with 4x, and 8x Nvidia L4 GPUS and the experiment was repeated each time.
Storage: The POD5 data resided on an attached EBS (Elastic Block Store) gp2 volume. To mitigate cold-start effects, each job was executed twice and only the second measurement was retained. This likely inflates apparent I/O throughput due to filesystem caching, but thats acceptable given that we are interested mainly in GPU compute scaling performance here.
Results
The performance metric is samples/second, as reported in the post-execution output of Dorado.
Multi GPU scaling

The cloud machine types used were the g6.2xlarge (single L4 GPU), g6.12xlarge (4x L4s) and g6.48xlarge (8x L4s). To browse machine types and prices in ResearchBox, just ask the assistant. e.g: "show me cloud machines with L4 NVIDIA GPUs"
How do CPU cores affect performance?
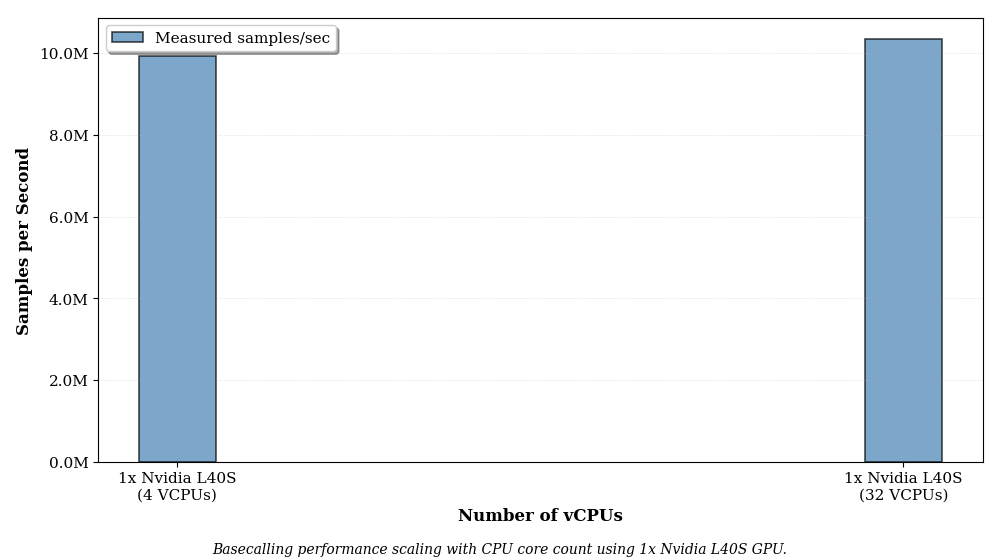
Dorado appears to max out a single CPU core. Going from 4 cores to 32 cores on a single Nvidia L40S GPU gives a negligable (~2%) speedup. Therefore you can use the lowest vcpu cloud instance type on AWS for each GPU type, with no loss of performance. This is in agreement with the 2023 perf benchmark by AWS using Dorado v0.3 : "Our cost evaluations revealed that the g5.xlarge instance delivers the lowest cost for basecalling a whole human genome (WHG) with the Guppy tool."
Comparing GPU architectures: Multi-GPU Turing vs Ada Lovelace
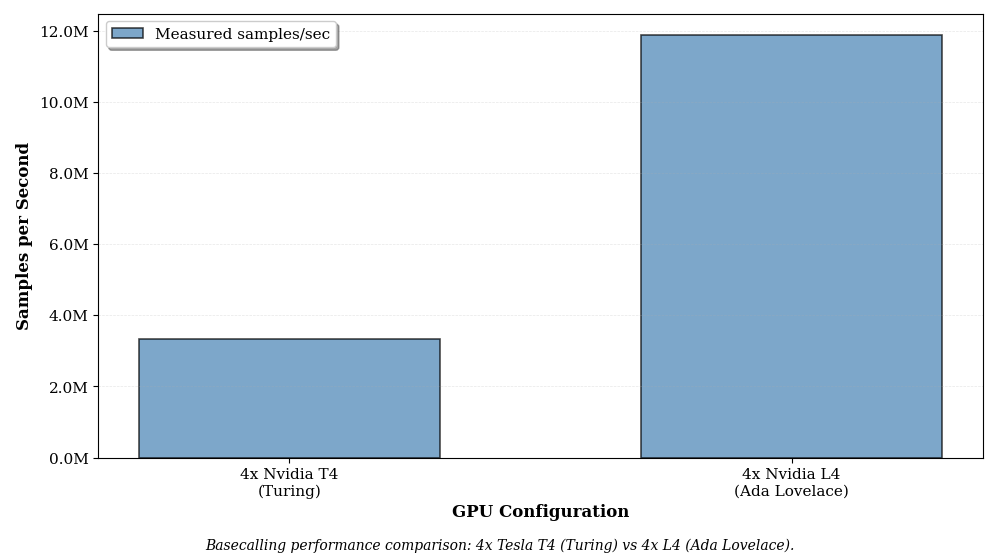
Basecalling performance on a 4x Nvidia T4 multi-GPU instance (g4dn.12xlarge) vs a 4x Nvidia L4 (g6.12xlarge)
Comparing GPU architectures: Single-GPU Ampere vs Ada Lovelace
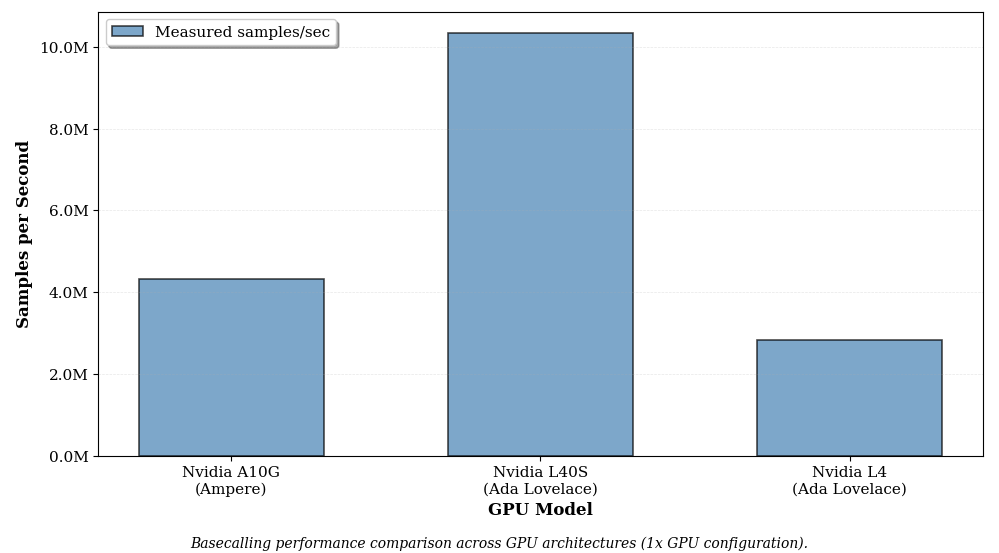
Basecalling performance on a single Nvidia A10G GPU (g5.8xlarge) vs a Nvidia L40S (g6e.8xlarge) vs a Nvidia L4 (g6.2xlarge). The g6.2xlarge has lower cpu count than the others but we have established earlier that cpu cores count does not materially change performance.
How to reproduce the linear scaling benchmark end-to-end on ResearchBox.ai in less than two hours
"create a researchbox for nanopore benchmarking"Optionally customize the cloud machine:
"show me the cheapest machine with a Nvidia L4 GPU"Select a machine type and name your new box. The assistant spins up a fully configured researchbox and streams it to your browser.
"how do i run the basecalling benchmark?"You can copy paste the commands into the linux terminal inside your Research Box. For v2 users, the assistant will offer to run the commands for you.
"resize my box"When asked to customize the cloud machine:
"show me all machines with more than one NVIDIA L4 GPU."Running time for the 1x, 4x, and 8x L4 insances is approximately 80 minutes, 20 minutes, and 10 minutes respectively.